Note
Go to the end to download the full example code.
RIXS calculations for an atomic model¶
Here we show how to compute RIXS for a single site atomic model with crystal field and electron-electron interactions. We take the case of Sr2YIrO6 from Ref. [1] as the material in question. The aim of this example is to illustrate the proceedure and to provide what we hope is useful advice. What is written is not meant to be a replacement for reading the docstrings of the functions, which can always be accessed on the edrixs website or by executing functions with ?? in IPython.
import edrixs
import numpy as np
import matplotlib.pyplot as plt
Specify active core and valence orbitals¶
Sr2YIrO6 has a \(5d^4\) electronic configuration and we want to calculate the \(L_3\) edge spectrum i.e. resonating with a \(2p_{3/2}\) core hole. We will start by including only the \(t_{2g}\) valance orbitals.
shell_name = ('t2g', 'p32')
v_noccu = 4
Slater parameters¶
Here we want to use Hund’s interaction \(J_H\) and spin orbit coupling \(\lambda\) as adjustable parameters to match experiment. We will take the core hole interaction parameter from the Hartree Fock numbers EDRIXS has in its database. These need to be converted and arranged into the order required by EDRIXS.
Ud = 2
JH = 0.25
lam = 0.42
F0_d, F2_d, F4_d = edrixs.UdJH_to_F0F2F4(Ud, JH)
info = edrixs.utils.get_atom_data('Ir', '5d', v_noccu, edge='L3')
G1_dp = info['slater_n'][5][1]
G3_dp = info['slater_n'][6][1]
F0_dp = edrixs.get_F0('dp', G1_dp, G3_dp)
F2_dp = info['slater_n'][4][1]
slater_i = [F0_d, F2_d, F4_d] # Fk for d
slater_n = [
F0_d, F2_d, F4_d, # Fk for d
F0_dp, F2_dp, # Fk for dp
G1_dp, G3_dp, # Gk for dp
0.0, 0.0 # Fk for p
]
slater = [slater_i, slater_n]
v_soc = (lam, lam)
Diagonalization¶
We obtain the ground and intermediate state eigenenergies and the transition
operators via matrix diagonalization. Note that the calculation does not know
the core hole energy, so we need to adjust the energy that the resonance will
appear at by hand. We know empirically that the resonance is at 11215 eV
and that putting four electrons into the valance band costs about
\(4 F^0_d\approx6\) eV. In this case
we are assuming a perfectly cubic crystal field, which we have already
implemented when we specified the use of the \(t_{2g}\) subshell only
so we do not need to pass an additional v_cfmat matrix.
edrixs >>> Running ED ...
Summary of Slater integrals:
------------------------------
Terms, Initial Hamiltonian, Intermediate Hamiltonian
F0_vv : 2.0000000000 2.0000000000
F2_vv : 2.1538461538 2.1538461538
F4_vv : 1.3461538462 1.3461538462
F0_vc : 0.0000000000 0.0881857143
F2_vc : 0.0000000000 1.0700000000
G1_vc : 0.0000000000 0.9570000000
G3_vc : 0.0000000000 0.5690000000
F0_cc : 0.0000000000 0.0000000000
F2_cc : 0.0000000000 0.0000000000
edrixs >>> Dimension of the initial Hamiltonian: 15
edrixs >>> Dimension of the intermediate Hamiltonian: 24
edrixs >>> Building Many-body Hamiltonians ...
edrixs >>> Done !
edrixs >>> Exact Diagonalization of Hamiltonians ...
edrixs >>> Done !
edrixs >>> ED Done !
Compute XAS¶
To calculate XAS we need to correctly specify the orientation of the x-rays
with respect to the sample. By default, the \(x, y, z\) coordinates
of the sample’s crystal field, will be aligned with our lab frame, passing
loc_axis to ed_1v1c_py can be used to specify a different
convention. The experimental geometry is specified following the angles
shown in Figure 1 of Y. Wang et al.,
Computer Physics Communications 243, 151-165 (2019). The default
setting has x-rays along \(z\) for \(\theta=\pi/2\) rad
and the x-ray beam along \(-x\) for
\(\theta=\phi=0\). Parameter scatter_axis can be passed to
xas_1v1c_py to specify a different geometry if desired.
Variable pol_type specifies a list of different x-ray
polarizations to calculate. Here we will use so-called \(\pi\)-polarization
where the x-rays are parallel to the plane spanned by the incident
beam and the sample \(z\)-axis.
EDRIXS represents the system’s ground state using a set of
low energy eigenstates weighted by Boltzmann thermal factors.
These eigenstates are specified by gs_list,
which is of the form \([0, 1, 2, 3, \dots]\). In this example, we
calculate these states as those that have non-negligible thermal
population. The function xas_1v1c_py assumes that the spectral
broadening is dominated by the inverse core hole lifetime gamma_c,
which is the Lorentzian half width at half maximum.
ominc = np.linspace(11200, 11230, 50)
temperature = 300 # in K
prob = edrixs.boltz_dist(eval_i, temperature)
gs_list = [n for n, prob in enumerate(prob) if prob > 1e-6]
thin = 30*np.pi/180
phi = 0
pol_type = [('linear', 0)]
xas = edrixs.xas_1v1c_py(
eval_i, eval_n, trans_op, ominc, gamma_c=info['gamma_c'],
thin=thin, phi=phi, pol_type=pol_type,
gs_list=gs_list)
edrixs >>> Running XAS ...
edrixs >>> XAS Done !
Compute RIXS¶
Calculating RIXS is overall similar to XAS, but with a few additional
considerations. The spectral width in the energy loss axis of RIXS it
not set by the core hole lifetime, but by either the final state lifetime
or the experimental resolution and is parameterized by gamma_f
– the Lorentzian half width at half maximum.
The angle and polarization of the emitted beam must also be specified, so
we pass pol_type_rixs to the function, which specifies the
includes the incoming and outgoing x-ray states. If, as is common in
experiments, the emitted polarization is not resolved
one needs to add both emitted polarization channels, which is what we will
do later on in this example.
eloss = np.linspace(-.5, 6, 400)
pol_type_rixs = [('linear', 0, 'linear', 0), ('linear', 0, 'linear', np.pi/2)]
thout = 60*np.pi/180
gamma_f = 0.02
rixs = edrixs.rixs_1v1c_py(
eval_i, eval_n, trans_op, ominc, eloss,
gamma_c=info['gamma_c'], gamma_f=gamma_f,
thin=thin, thout=thout, phi=phi,
pol_type=pol_type_rixs, gs_list=gs_list,
temperature=temperature
)
edrixs >>> Running RIXS ...
edrixs >>> RIXS Done !
The array xas will have shape
(len(ominc_xas), len(pol_type))
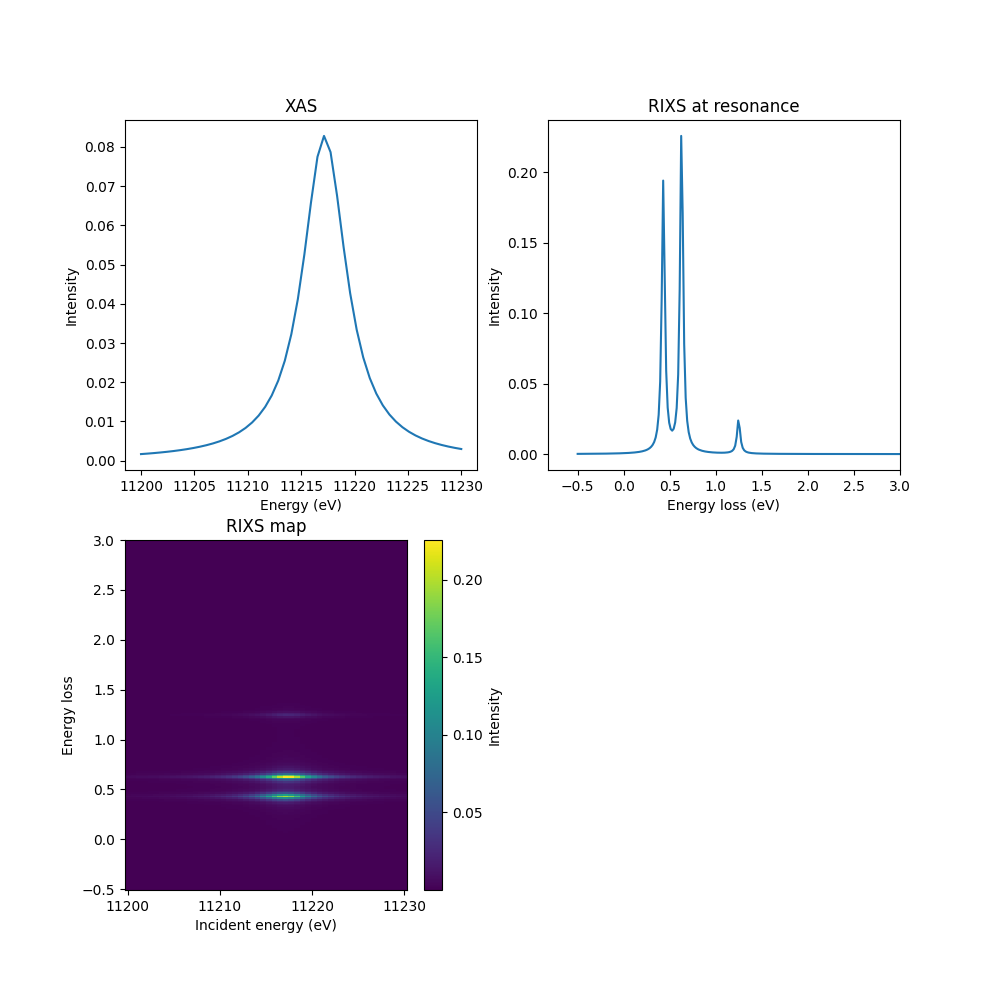
Plot XAS and RIXS¶
Let’s plot everything. We will use a function so we can reuse the code later.
Note that the rixs array rixs has shape
(len(ominc_xas), len(ominc_xas), len(pol_type)). We will use some numpy
tricks to sum over the two different emitted polarizations.
fig, axs = plt.subplots(2, 2, figsize=(10, 10))
def plot_it(axs, ominc, xas, eloss, rixscut, rixsmap=None, label=None):
axs[0].plot(ominc, xas[:, 0], label=label)
axs[0].set_xlabel('Energy (eV)')
axs[0].set_ylabel('Intensity')
axs[0].set_title('XAS')
axs[1].plot(eloss, rixscut, label=f"{label}")
axs[1].set_xlabel('Energy loss (eV)')
axs[1].set_ylabel('Intensity')
axs[1].set_title(f'RIXS at resonance')
if rixsmap is not None:
art = axs[2].pcolormesh(ominc, eloss, rixsmap.T, shading='auto')
plt.colorbar(art, ax=axs[2], label='Intensity')
axs[2].set_xlabel('Incident energy (eV)')
axs[2].set_ylabel('Energy loss')
axs[2].set_title('RIXS map')
rixs_pol_sum = rixs.sum(-1)
cut_index = np.argmax(rixs_pol_sum[:, eloss < 2].sum(1))
rixscut = rixs_pol_sum[cut_index]
plot_it(axs.ravel(), ominc, xas, eloss, rixscut, rixsmap=rixs_pol_sum)
axs[0, 1].set_xlim(right=3)
axs[1, 0].set_ylim(top=3)
axs[1, 1].remove()
plt.show()
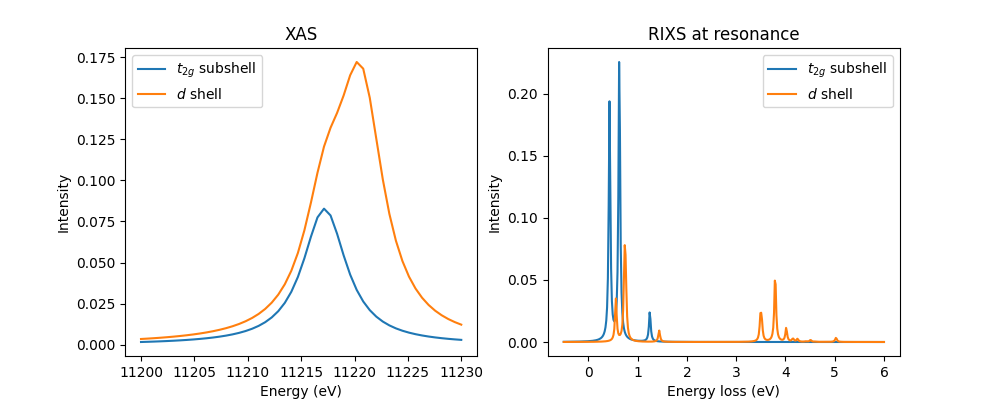
Full d shell calculation¶
Some researchers have questioned the appropriateness of only including the
\(t_{2g}\) subshell for iridates [2]. Let’s test this. We specify that
the full \(d\) shell should be used and apply cubic crystal field matrix
v_cfmat. We shift the energy offset by \(\frac{2}{5}10D_q\), which
is the amount the crystal field moves the \(t_{2g}\) subshell.
ten_dq = 3.5
v_cfmat = edrixs.cf_cubic_d(ten_dq)
off = 11215 - 6 + ten_dq*2/5
out = edrixs.ed_1v1c_py(('d', 'p32'), shell_level=(0, -off), v_soc=v_soc,
v_cfmat=v_cfmat,
c_soc=info['c_soc'], v_noccu=v_noccu, slater=slater)
eval_i, eval_n, trans_op = out
xas_full_d_shell = edrixs.xas_1v1c_py(
eval_i, eval_n, trans_op, ominc, gamma_c=info['gamma_c'],
thin=thin, phi=phi, pol_type=pol_type,
gs_list=gs_list)
rixs_full_d_shell = edrixs.rixs_1v1c_py(
eval_i, eval_n, trans_op, np.array([11215]), eloss,
gamma_c=info['gamma_c'], gamma_f=gamma_f,
thin=thin, thout=thout, phi=phi,
pol_type=pol_type_rixs, gs_list=gs_list,
temperature=temperature)
fig, axs = plt.subplots(1, 2, figsize=(10, 4))
plot_it(axs, ominc, xas, eloss, rixscut, label='$t_{2g}$ subshell')
rixscut = rixs_full_d_shell.sum((0, -1))
plot_it(axs, ominc, xas_full_d_shell, eloss, rixscut, label='$d$ shell')
axs[0].legend()
axs[1].legend()
plt.show()
edrixs >>> Running ED ...
Summary of Slater integrals:
------------------------------
Terms, Initial Hamiltonian, Intermediate Hamiltonian
F0_vv : 2.0000000000 2.0000000000
F2_vv : 2.1538461538 2.1538461538
F4_vv : 1.3461538462 1.3461538462
F0_vc : 0.0000000000 0.0881857143
F2_vc : 0.0000000000 1.0700000000
G1_vc : 0.0000000000 0.9570000000
G3_vc : 0.0000000000 0.5690000000
F0_cc : 0.0000000000 0.0000000000
F2_cc : 0.0000000000 0.0000000000
edrixs >>> Dimension of the initial Hamiltonian: 210
edrixs >>> Dimension of the intermediate Hamiltonian: 1008
edrixs >>> Building Many-body Hamiltonians ...
edrixs >>> Done !
edrixs >>> Exact Diagonalization of Hamiltonians ...
edrixs >>> Done !
edrixs >>> ED Done !
edrixs >>> Running XAS ...
edrixs >>> XAS Done !
edrixs >>> Running RIXS ...
edrixs >>> RIXS Done !
As expected, we see the appearance of excitations on the energy scale of \(10D_q\) in the XAS and RIXS. The low energy manifold is qualitatively, but not quantiatively similar. This makes it clear that the parameterization of Sr2YIrO6 is dependent on the model.
Footnotes
Total running time of the script: (0 minutes 3.985 seconds)